Cross-validation settings#
Cross-validation is a fundamental core of machine learning analysis because it facilitates out-of-sample generalization and parameter tuning (see wiki. NeuroMiner has been built to flexibly create custom cross-validation schemes depending on your data problem. NeuroMiner has been built around performing repeated, nested cross-validation, which is a very robust method that increases the likelihood of generalization.
The cross-validation works by first defining the following settings:
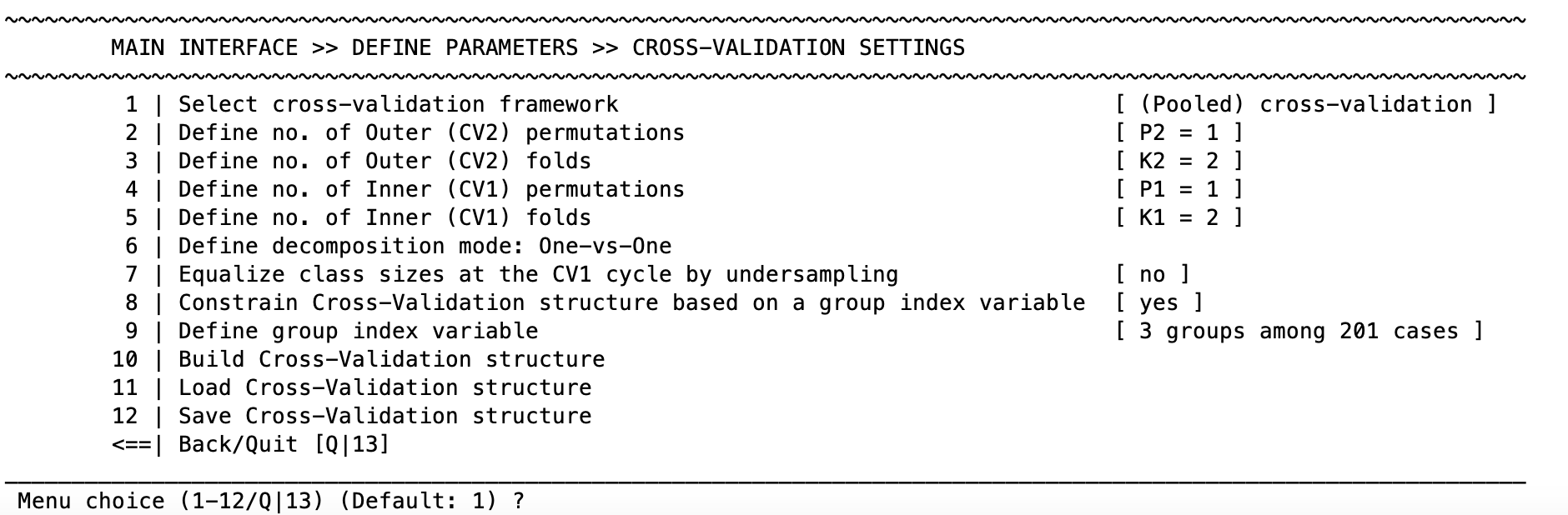
Fig. 7 NeuroMiner cross-validation framework setup#
Select cross-validation framework#
This option allows the user to select either k-fold cross-validation scheme (including leave-one-out) or a leave-group-out analysis with the following options:
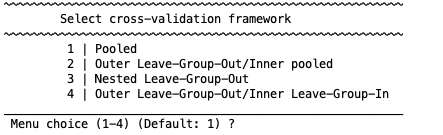
Fig. 8 NeuroMiner cross-validation frameworks#
NeuroMiner has been built around the gold standard of repeated, nested cross-validation (Filzmoser et al., 2009) and therefore the default options are reflective of this. For a description of cross-validation, you could check-out the supplementary material of this publication: Koutsouleris et al., 2016. In brief, leave-one-out cross-validation and k-fold cross-validation risk overfitting (i.e., your models not generalizing past your sample) in the context of (hyper)parameter optimisation (e.g., optimizing C parameters) or the optimization of feature selection based on the predictive accuracy on the held-out dataset (e.g., using filters or wrappers discussed in Ensemble generation strategies). Nested cross-validation mitigates against overfitting and provides models that are more likely to generalize – which is the central aim of machine learning and science generally.
Nested cross-validation in NeuroMiner#
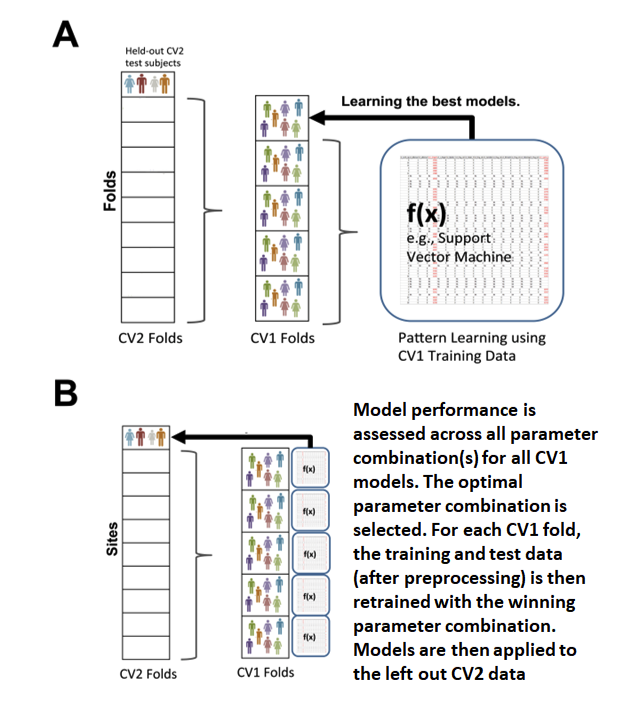
Fig. 9 Nested cross-validation#
A depiction of nested cross-validation is represented in Fig. 9. In NeuroMiner the outer cross-validation cycle of a nested cross-validation scheme is designated as CV2 and the inner cross-validation folds are designated as CV1. Models are trained in the CV1 cycle and then the best performing models are applied to the CV2 data. This separation of CV2 and CV1 data avoids overfitting.
First the data is split into k_outer folds in the outer CV2 set. One of these CV2 folds is held out and serves as the test set in this round. The rest of the data goes into the ’nest’ where it is again subdivided into k_inner folds (note: k_inner does not have to be = to k_outer). In the ‘nest’, an inner test-fold is held out, and the rest of the data is used for training. Models associated with each parameter combination are created in the CV1 (inner) training data and applied to the CV1 test data (as shown in Fig. 9, A). The accuracy is assessed across all parameter combinations for all CV1 models. The optimal parameter combination is chosen based on a criteria set by the user. For each CV1 fold, a model is retrained on the processed training and test data with the winning parameter or parameter combination. The models are then applied to the held-out CV2 test data (as shown in Fig. 9, B).
In cases where there are many (hyper)parameter combinations or when a variable selection procedure is employed, then the user can choose to keep a percentage of top performing models for each fold, instead of only the single optimum model. This is further discussed in Learning algorithm parameters.
Let’s look at the other cross-validation frameworks included in NeuroMiner:
Cross-validation frameworks in NeuroMiner
1 | Pooled Outer and inner cross-validation folds will be automatically and randomly defined.
2 | Outer Leave-Group Out/Inner pooled The outer CV2 folds will be defined by the user using a vector defining groups (e.g., if there are different sites then this will test the ability of the classifiers that are trained with pooled site data to generalise to new sites).
3 | Nested Leave-Group-Out Both the outer and inner cycles will be separated based on the grouping vector that the user enters (e.g., in the site example, this will mean that the models will be optimised to generalise across sites in the CV1 cycle and then applied to different sites in the CV2 cycle).
4 | Outer Leave-Group-Out/Inner Leave-Group-In The models in the inner CV1 cycle will only be trained in one group prior to being applied to all other groups (e.g., this will test the robustness of models from single sites). See Fig. 9 for more details and imagine that the folds are sites or diagnoses or any other grouping variable.
Repeated cross-valiation#
To further avoid overfitting and increase the generalizability of the model predictions, NeuroMiner also allows the user to implement ’repeated’ cross-validation for each of the CV cycles. This involves shuffling the subjects prior to the definition of the folds so that there are different subjects in the test/training folds. This is usually done for both the CV2 and CV1 cycles.
A clear depiction of how repeated cross-validation works is difficult, but it is important to keep in mind that there will be a lot of models produced under these schemes; e.g., if you have a standard 10x10 CV1/CV2 framework and you’re choosing the optimal model for each fold, then you’ll have 10,000 models. A way to understand how these are applied to an individual subject is to consider that all models in which that subject is not included in the CV1 training/testing are applied to them – because if they were included then this would be information leakage. On the surface, having 10,000 models is not so great for the interpretation or understanding of a phenomenon under investigation. However, doing this can be really good for generalization when it’s unlikely that there is a singular model that can be found and we need something that works in practice – e.g., for many complex problems in psychiatry. For an interesting discussion on this topic, see Breiman et al., 2001.
Note
Repetition is only available for some cross-validation frameworks (when it makes sense).
Define the number of folds & permutations in CV1 & CV2#
In the cross-validation menu (Fig. 7) you can define the number of permutations (“repeats”) and folds for CV1 and CV2 respectively using the options 2 to 5 (note that these might change depending on the cross-validation framework you choose!).
Choosing the number of permutations#
For some cross-validation frameworks, you can define the number of repetitions that you of the cross-validaiton procedure. If you choose to use repeated cross-validation, the subjects are shuffled and new folds are created. To choose the number of permutations, try to find a balance between the computational time that your analysis is taking and the stability of the predictions from your modeling.
Choosing the number of folds#
The number of folds is usually based on the number of subjects you have and what kind of analyses you want to compute.
For an analytic example of when the number of CV1 folds is crucial for the outcome see the section on Ensemble generation strategies – e.g., when using wrappers that are optimizing features based on the subjects in the CV1 test folds.
Tip
If you enter “-1” as the number of CV1 or CV2 folds, then NeuroMiner implements a leave-one-out cross-validation and the permutations option is not possible (because it doesn’t matter if you shuffle the data if every single subject is left out). The number of CV folds is usually based on the number of subjects that you have.
Equalize class sizes at the CV1 cycle by undersampling#
Unbalanced group sizes (e.g., less people transitioning to illness than people who do not) pose a problem for many machine learning techniques. Usually, the trained model shows a bias towards the majority group and you’ll get unbalanced predictions (e.g., your specificity will be high, but sensitivity low).
In NeuroMiner there are three ways to deal with this:
Undersampling the majority group during training
Weighting of hyperplanes in case of margin-based classifiers (SVM as implemented in LIBSVM or LIBLINEAR)
Oversampling using “Adaptive synthetic sampling” (ADASYN, see He et al., 2008) which is recommended for all other algorithms in NM
In the classification setting, option 1 allows the user to undersample the majority group so that the groups are balanced when the models are created in the CV1 folds. In the case of the regression framework, undersampling aims at creating a uniform distribution of the target label(s).
After selecting the undersampling option (for classification, it’s called “Equalize class sizes at the CV1 cycle by undersampling”; for regression, it’s “Equalize label histogram at the CV1 cycle by undersampling“), new settings will be added to your menu:
In the case of classification, selecting the menu will now include:
Define positive/negative ratio after equalization (1=>same amount of positive/negative cases) [ 1.5 ]
Shuffle removed observations to CV1 test data [ yes ]
The target ratio is the ratio of the bigger to smaller class sample sizes. Equal class sizes can be selected by entering in ”1”. The default is 1.5 because otherwise the models may not generalize well.
The menu entry “Shuffle removed observations to CV1 test data” gives the user the option to add the excluded CV1 subjects in the training folds to the CV1 test folds, e.g. for identifying optimal hyperparameters for model generalization.
In the case of regression, the menu will now include:
1 | Define target label(s) for uniform distribution
2 | Define minimum # of observations at the lower end of label histogram
3 | Define minimum # of observations at the upper end of label histogram
4 | Define # of bins in label histogram
5 | Show original histogram of the entire data
6 | Show (equalized) histogram of CV1 training partition [1,1] (build CV structure first!)
7 | Define positive/negative ratio after equalization (1=>same amount of positive/negative cases): This is the ratio of the bigger to smaller class sample sizes. Equal class sizes can be selected by entering in ”1”. The default is “1.5” because otherwise the models may not generalize well.
8 | Shuffle removed observations to CV1 test data: The next option gives the user the option to add the excluded CV1 subjects in the training folds to the CV1 test folds. This option is can be important to judge generalizability of the models to the CV2 folds (and ultimately to real-life).
Note
Small‑batch safeguards (NM 1.4)
To stabilize stratified CV with rare strata:
Remove n=1 batches from CV1 training partitions (guard against singleton strata).
Move batches with N ≤ 5 to test folds to avoid leakage and optimistic training.
Logging: NM writes a CV cycle generation log listing fold membership, removed singletons, and relocated batches for reproducibility.
Enable under: Cross‑validation settings → Stratification options → Small‑batch handling.
Multiclass classification setting: Define decomposition mode#
1 | One-vs-One
2 | One-vs-All
This option only shows in case your label is a multiclass classification label. You have to define the decomposition mode you want to use.
One vs. One means that pairwise classification models will be fit between all possible group pairs. If you have a label containing three groups, the following binary three models will be trained: group 1 vs. 2, group 1 vs. 3, group 2 vs. 3.
One vs. All means that for each of the groups in your label, a model will be fit to classify this group from all other groups. If you have a label containing three groups, the following models will be trained: group 1 vs. (2+3), 2 vs. (1+3), 3 vs. (1+2).
Constrain Cross-Validation structure based on a group index variable#
You can enter a vector defining groups which will be used additionally to the label to stratify the cross validation folds, i.e., the function will make sure to distribute the groups equally across folds.
Build, load & save the cross-validation structure#
Build CV2/CV1 structure Once the options above have been set, the user needs to build the CV2/CV1 structure. This literally creates all the folds and indexes the subjects that are to be included in the folds. These can be found in the NM structure (i.e., in “NM.cv”).
Important
The user must select Build the cross-validation structure before exiting the cross-validation menu. Otherwise, the cross-validation framework will not be saved within the NM structure.
Load CV structure If you have saved the CV structure for your data with the subjects indexed into each fold/permutation, you can load it using this function. This can be useful if you establish a new analysis with new settings, but you want to load the CV structure that you created before.
Save CV structure This gives the user the option to save the indexed CV structure containing the folds and permutations for later use. It is saved in a .mat file.
