Installation & Configuration#
Compatibility#
NeuroMiner has been tested using Windows, Linux (Cent OS), and Mac OS operating systems and works with MATLAB2020b and above. However, to make use of some advanced functionalities which resort to MATLAB’s App Designer (i.e., sample size simulation, MLI viewer, ROI mapper) and MATLAB’s more recent Python integration functions, you need MATLAB R2021b or above.
We recommend the ”Statistics and Machine Learning” and the ”Optimization” toolboxes in order to use advanced training options in the matLearn toolbox and for imputation functions.
For non-supported operating systems, NeuroMiner has a number of programs installed that may require independent compilation using a C compiler (e.g., gcc for Linux or Xcode for Mac).
Important
If you want to use NeuroMiner on macOS, see Setting up NeuroMiner on macOS.
For some functionalities, NeuroMiner resorts to recognized Python libraries. Thus, a Python installation (tested with Python 3.8) may be required. If you have never called a Python script from MATLAB, you initially need to set the correct Python environment in MATLAB. You can find instructions on how to set the Python version in the MATLAB Help Center. You only have to configure the path once.
To install the necessary Python packages, run the following line in the command line.
pip install -r Python/requirements.txt
Alternatively, you can install them separately, the functionalities require the following:
We recommend that you set up a separate virtual environment for this so that these packages do not interfere with your other installations.
Downloading and Initializing NeuroMiner#
You can download NeuroMiner from the GitHub repository by clicking on the green Code button on the right-hand side and then choosing the Download ZIP option (see Fig. 1).
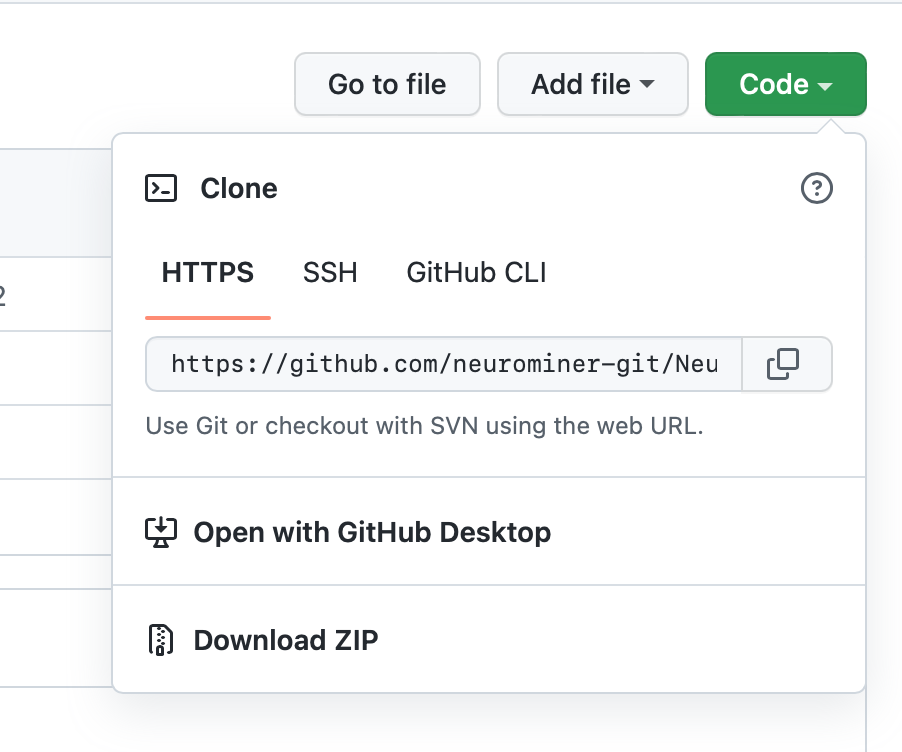
Fig. 1 Installation by downloading the NeuroMiner_1.2 ZIP-Folder from GitHub#
Note
Alternatively, you can download NeuroMiner from the command line:
Go to the directory on your disk where you want to save it by typing
cd PATH_TO_DIRECTORY_OF_CHOICE
Then, to download the software, type
git clone https://github.com/neurominer-git/NeuroMiner_1.2.git
NeuroMiner works in two main modes: matrix and neuroimaging (i.e., NIFTI or FreeSurfer files). For neuroimaging using NIFTI data format, it is necessary to have a working copy of the Statistical Parametric Mapping (SPM) toolbox installed. We also recommend downloading the WFU Pickatlas toolbox to take advantage of this selection tool when inputting data. For surface-based analyses, it is necessary to have a FreeSurfer distribution installed.
Once the program is downloaded, it needs to be added to the MATLAB path by using the addpath command from the command line or by using the “Set Path” button on the toolbar (e.g., “addpath /home/NM/NeuroMiner”). NeuroMiner can then be launched by typing nm on the command line. To load the program faster, you can type nm nosplash and to enter expert mode type nm nosplash expert. The first time that NeuroMiner is run, a file selector box will appear first asking to define the SPM directory and then the FreeSurfer directory. If these directories are not available, then simply press cancel and NeuroMiner will be launched in non-imaging mode–i.e., a mode that restricts options only to matrix data. Otherwise, NeuroMiner will be launched in matrix and neuroimaging mode (Fig. 1).
Fig. 2 Display screen of NeuroMiner in different modes. NeuroMiner will be loaded in non-imaging mode (upper) when the SPM or FreeSurfer directories are not provided during start-up (upper). If the directories have been provided, it will be started in standard imaging and matrix data mode (lower).#
Caution
Once NeuroMiner is initialized and added to the path, there can be incompatibilities between some MATLAB functions on some systems. For example, there can be a incompatibility with the function ttest2. In these cases, NeuroMiner needs to be removed from the path in order to use the original function.
