Ensemble generation strategies#
NeuroMiner automatically works with ensemble learning at the CV2 level across the CV1 folds of a nested, repeated cross-validation design (see Fig. 9), but it also includes the ability to apply filters and wrappers that optimize predictions by selecting features within these folds (for more information, see wiki and Kolhavi & John.
Filter-based ensemble generalization#
NeuroMiner has two methods of applying filters:
As part of the preprocessing of features by pre-selecting variables based on their relationship to the target (rank/ weight features)
As an ensemble method where different variable sets (variable subspaces) can be chosen based on the performance of the machine learning training algorithm.
The latter case is known as constrained feature optimization because the variable sets are produced based on the performance of an algorithm (e.g., correlation), the sets are evaluated with the algorithm, and then the models corresponding to well-performing variable sets are chosen as an ensemble.
The first step is to turn the option specifying the ”Train filter methods on CV1 partitions” to “yes”. This will generate the following menu:
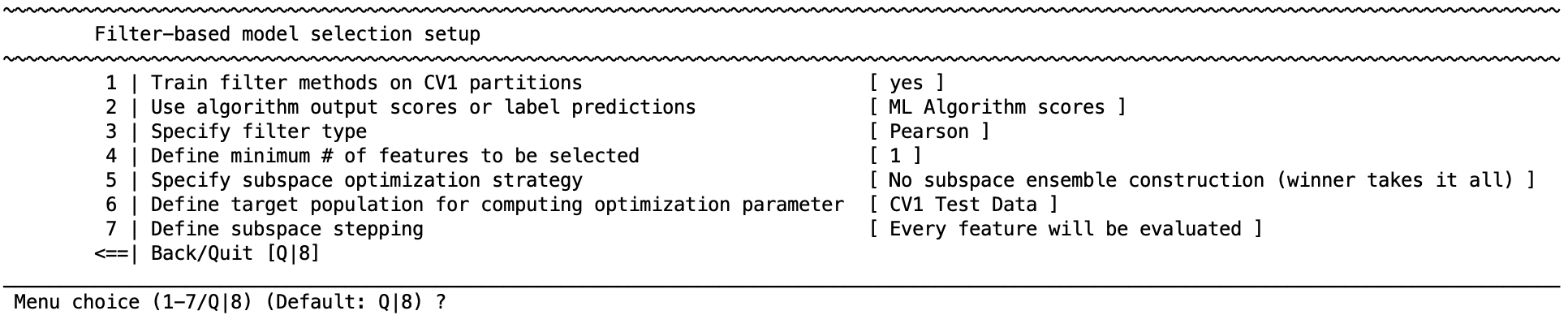
1 | Train filter methods on CV1 partitions#
Turn constrained feature optimization filtering on or off.
2 | Use algorithm output scores or label predictions#
Choose the feature sets based on majority voting of the predicted labels (hard decision ensemble) or based on an average of the predicted probabilities from the machine learning algorithm (soft decision ensemble).
3 | Specify filter type#
Specify what filter will be used to obtain the feature sets. These methods are the same as in the preprocessing filter.
4 | Define minimum number of features to be selected#
This gives the option to define a minimum number of features that are retained when choosing feature sets.
5 | Specify subspace optimization strategy#
Once the subspaces are defined and the models are evaluated, this section will define which subspaces/models are retained. There is the option to retain one model of the best performing subspace, or an ensemble of models across well-performing subspaces.
1 | Subspace with maximum argmax: Argmax (arguments of the maxima) are the points of the domain of some function at which the function values are maximized. This option picks the maximum argmax across the feature subspaces (winner-takes-it-all).
2 | Subspace ensemble with maximum argmax: Picks the X most predictive subspaces with reference to the argmax.
3 | Subspace ensemble with maximum argmax above a percentile Picks the top X% of subspaces.
4 | All-subspace ensemble: Uses all subspaces when training the model.
Provided that an ensemble-based method is selected as the subspace selection strategy (options 2-4), an additional menu entry (2 | Define ensemble optimization method) allows you to choose the ensemble optimization method from one of the following, essentially referring to the way in which you want the base models generated for each feature subspace to be combined:

1 | Simply aggregate all learners into ensemble: This is also known as bagging and consists of averaging across the base learners trained independently in parallel.
2 | Optimize ensemble using backward base learner elimination: This uses a greedy search algorithm in order to find an optimal subset of base learners by backward search, such that all base learners are initially included and then learners are recursively removed in order to achieve an optimal solution. The optimization can be done based on different criteria (default: Entropy & Performance, Entropy of ensemble’s component models, Kappa-Diversity among ensemble’s component decisions or Bias-Variance decomposition of ensemble’s component errors). Either the algorithm output scores (soft ensemble) or the label predictions (hard ensemble) can be used in order to construct the ensemble. Moreover, you can specify a minimum number of base learners to be included in the ensemble and enable weigthing of the base learners based on the inverse of their error. This opens a sub-menu to further specify additional parameters:

3 | Optimize ensemble using forward base learner selection: This uses a greedy search algorithm in order to find an optimal subset of base learners by forward search, such that initially only one base learner is included and then learners are recursively added in order to achieve an optimal solution. The other settings are similar to those described for the backward base learner elimination.
4 | Create a single classifier using probabilistic feature subspace construction: This option selects the optimal feature subspaces based on the agreement across the different base learners for each specific feature. For this, you need to specify a threshold for the percentage of times that a feature is selected across CV1 feature subspaces and only the features above this threshold will be used to build the ensemble classifier.
5 | Use AdaBoost to determine optimal weighting of base learners: Here, the popular boosting algorithm AdaBoost is used in order to compute a weighted majority vote of the weak hypotheses generated by the weak learners. More details on how the AdaBoost algorithm works can be found in Freund, Y., & Schapire, R. E. (1997) or Schapire, R. E. (2013).
6 | Define target population for computing optimization parameter: You can choose the best performing models using the CV1 training data, the CV1 test data, or the CV1 training & the test data. This means that the subspaces will be selected on the basis of how they predict the labels in each of these data folds. Selection based on training and test data is recommended.
7 | Define subspace stepping: This option defines how the subspaces are formed. The features are ranked based on the association between them and the target variable, and then they are divided into subspaces based on blocks of X% of features; e.g., blocks of 10% of features would divide the data into the top 10% performing features, then the top 20% of features, then the top 30% of features, and so on.
Important
It is important to note that once the feature subspaces are defined, the features within the winning subspaces can then be used in a wrapper to further optimize performance.
6 | Define target population for computing optimization parameter#
You can choose the best performing models using the CV1 training data, the CV1 test data, or the CV1 training & the test data. This means that the subspaces will be selected on the basis of how they predict the labels in each of these data folds. Selection based on training and test data is recommended.
7 | Define subspace stepping#
This option defines how the subspaces are formed. The features are ranked based on the association between them and the target variable, and then they are divided into subspaces based on blocks of X% of features; e.g., blocks of 10% of features would divide the data into the top 10% performing features, then the top 20% of features, then the top 30% of features, and so on. It is important to note that once the feature subspaces are defined, the features within the winning subspaces can then be used in a wrapper to further optimize performance.
Wrapper-based model selection#
The wrapper methods in NeuroMiner use either Greedy feature selection or simulated annealing to select feature combinations that maximize the predictive accuracy of a model in the CV1 data. You will see the following menu:
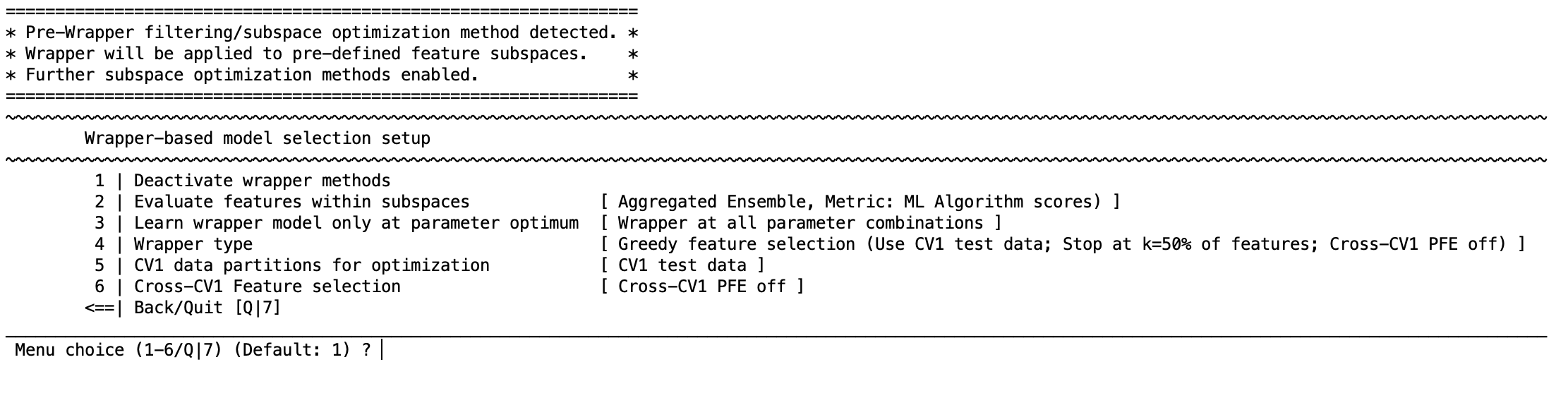
4 | Wrapper type#
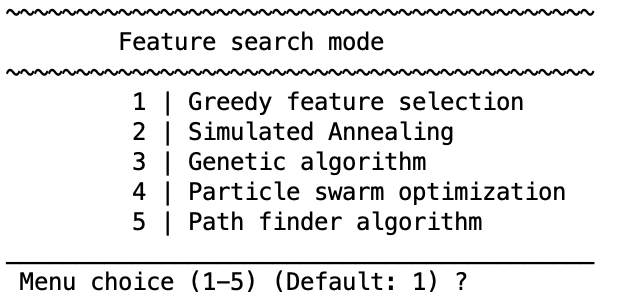
Select greedy feature selection or simulated annealing.
Greedy feature selection#
1 | Search direction [ Forward ]
2 | Early stopping [ Stop at 50% of feature pool ]
3 | Feature stepping [ Each feature will be evaluated ]
4 | Kneepoint-based threshold detection [ disabled ]
Greedy feature selection aims to find the best subset of features by iteratively adding/ excluding features. This means that at each step, the algorithm chooses the features that improve the performance at this stage, i.e., the algorithm makes a local optimal choice and may, thus, not end up at a global optimum.
The “search direction” must be provided (forward or backward), which determines whether an empty subspace is filled with features that are the most predictive of the target variable (forward selection) or whether all the variables are entered into the subspace and then variables are removed until the optimal model is found (backward).
If you enable “early stopping”, a defined percentage of variables is remaining in the feature pool, i.e., when this percentage is reached no more variables are added to the analysis. This setting is useful when you want to find parsimonious solutions and restricting the features to be included can lead to better solutions.
“Feature stepping” can also be changed from adding/removing single features and then testing the model, or adding/removing percentages of the remaining features before testing the model. Adding percentages (e.g., 5%) normally results in a faster processing time.
Simulated annealing#
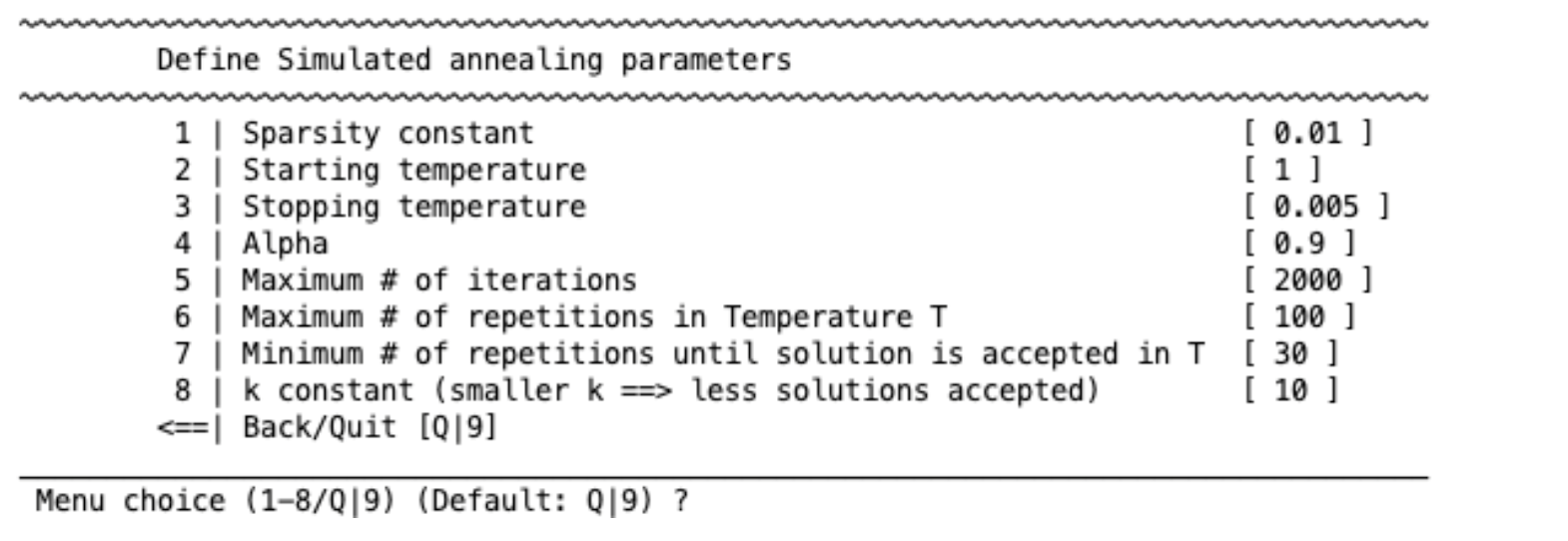
Simulated annealing is a probabilistic technique for approximating the global maximum of a function. The settings are standard parameters for any simulated annealing analysis.
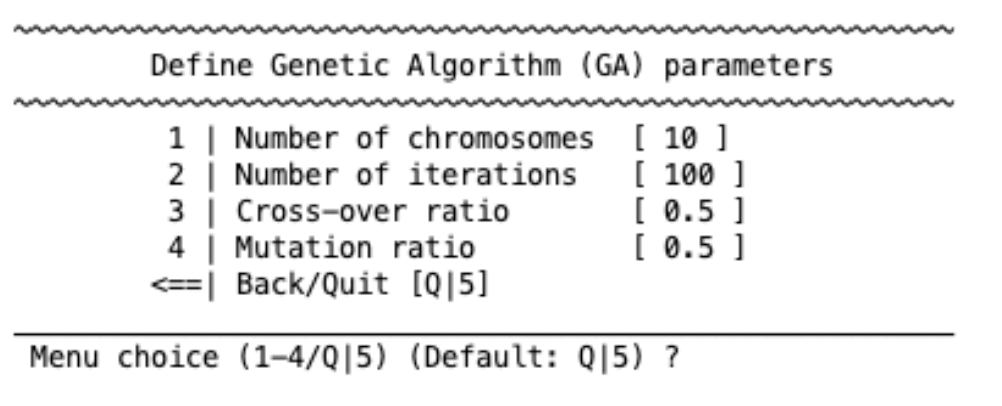
Genetic algorithm#
A genetic algorithm tries to find the best feature selection method following strategies similar to natural selection in evolution. You can specify the following parameters (see the linked paper for more details):
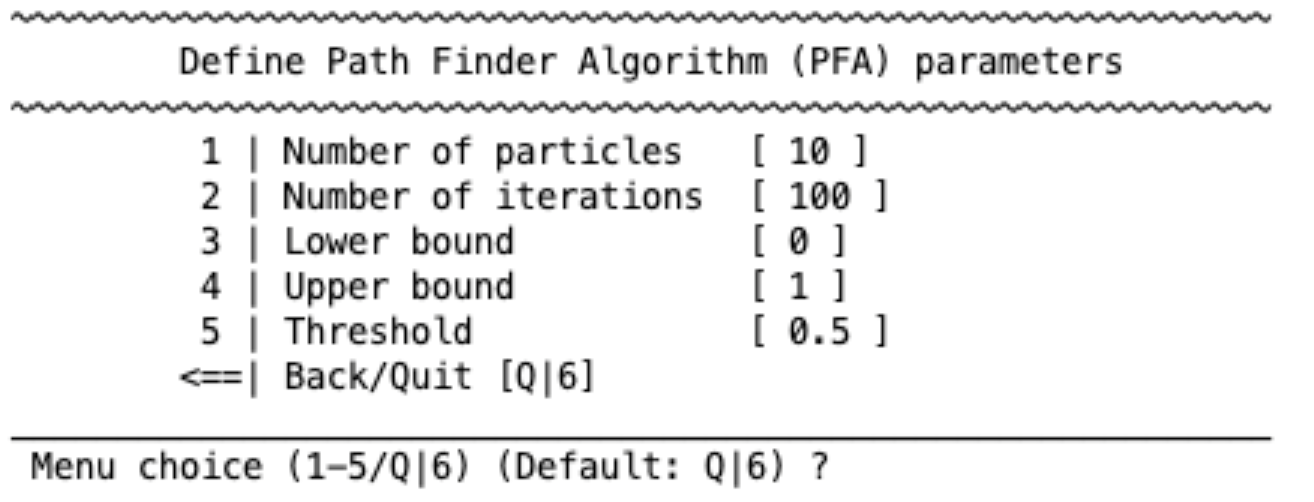
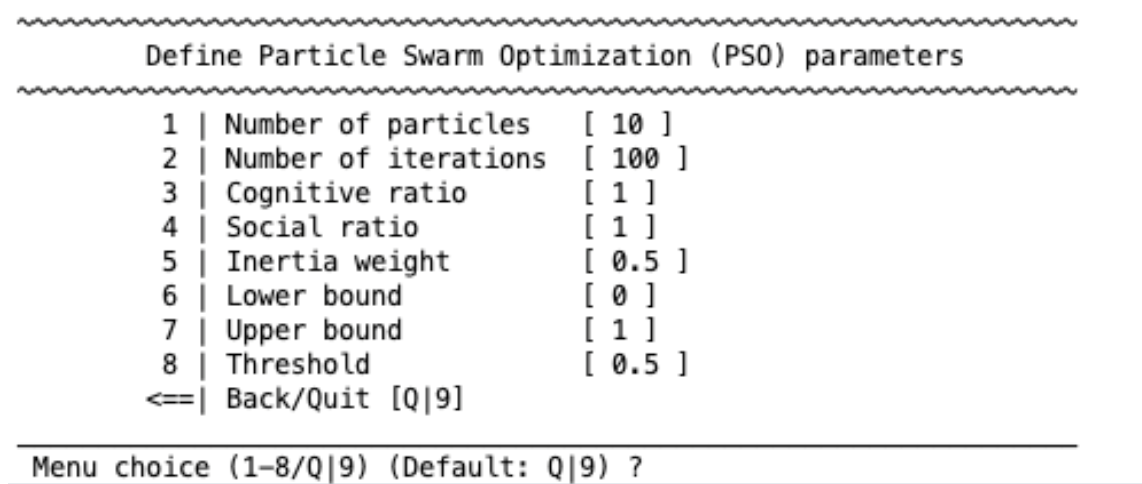
Particle swarm optimization#
particle swarm optimization algorithm is based on the behavior of birds. It has the following parameters:
5 | CV1 data partitions for optimization#
Choose either the CV1 training, test, or test & training data to optimize the feature set.
6 | Cross-CV1 feature selection#
In a nested cross-validation design, you will have a selection of features for each of the CV1 partitions. This step allows you to select features across the CV1 folds and will reveal another menu:
1 | Optimize feature selection across CV1 partitions [ yes ]
2 | Probabilistic feature extraction mode [% Cross-CV1 feature selection agreement ]
3 | Apply consistency-based ranking to [ Prune unselected features from each model and retrain models ]
Probabilistic feature extraction#
1 | Cross-CV1 feature selection agreement with tolerance window
2 | Absolute number of most consistently selected features
3 | Percentage of most consistently selected features
Cross-CV1 feature selection agreement with tolerance window This option allows you to select features that occur across CV1 partitions at a certain percentage rate. For example, select only features that appear across CV1 folds 75% of the time or more. Sometimes the threshold that is set does not return any features, and therefore you will also be asked to define a tolerance value for this circumstance. For example, if the 75% criterion is not met, then reduce this by 25% (i.e., so then you are effectively selecting features 50% of the time). You will also be asked to define a minimum number of features that must be selected each time (e.g., one feature).
Absolute number of most consistently selected features This option orders the features based on the amount of times they were selected across the CV1 folds. Then you can select the number of features occurring at the top of the list (e.g., you could select the top 10 most consistently selected features).
Percentage of most consistently selected features This option sorts in the same way as described for absolute number of consistently selected features and then establishes a cut-off. The features above this cut-off are kept. For example, if 90% is the cut-off then it will select the top 10% of the most consistently selected features across the CV1 folds.
Apply consistency-based ranking#
1 | Retrain all CV1 models after pruning them from unselected features
2 | Retrain all CV1 models using the same selected feature space
Once the features are selected, you can either ”1 | Retrain all CV1 models” after pruning the unselected features. Or you can completely retrain the models using the single optimized feature set that was found in the previous steps.